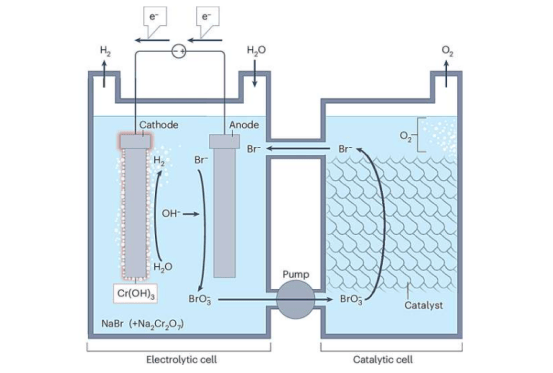
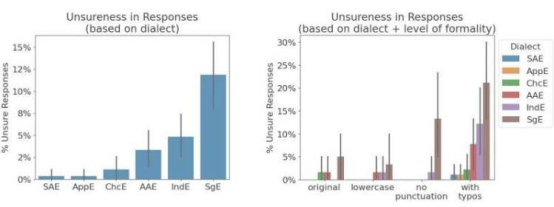
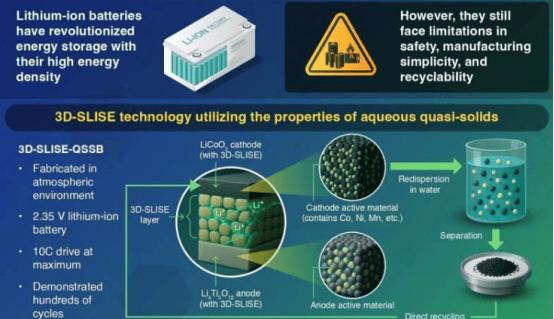
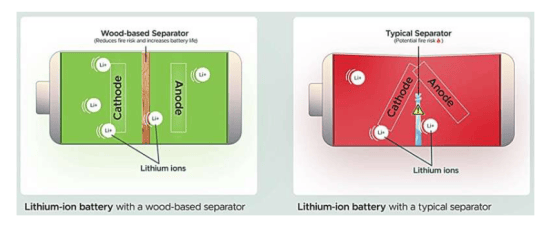
A research team from the University of Cologne has recently developed a new method called DynaTag, which efficiently identifies binding sites of transcription factors to DNA. This technology overcomes the limitations of existing methods, achieving high-precision detection in low-input samples and at the single-cell level. The related research findings have been published in the journal Nature Communications.
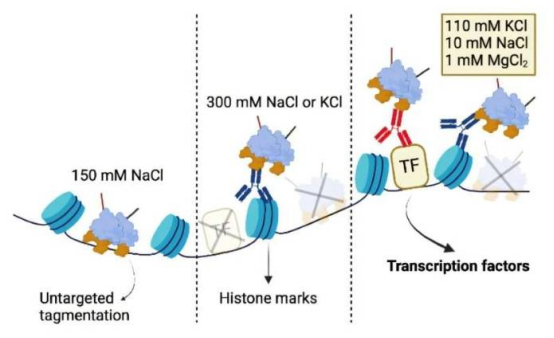
Transcription factors are proteins that bind to DNA and regulate gene expression, but their dynamic interactions pose challenges for precise mapping. Traditional methods such as ChIP-seq and CUT&RUN have deficiencies in sensitivity and resolution. Dr. Robert Hänsel-Hertsch, Principal Investigator at the Center for Molecular Medicine Cologne, stated: "DynaTag outperforms existing technologies in sensitivity and result resolution, enabling more precise mapping of DNA regions where transcription factors bind."
The research team validated the practicality of DynaTag using mouse models. In experiments with small cell lung cancer, the method successfully identified transcription factors activated after chemotherapy, which may be associated with tumor drug resistance and metastasis. Hänsel-Hertsch noted: "DynaTag has helped us discover transcription factors that regulate key signaling pathways, providing new insights into understanding tumor progression."
This technology can also be applied to the analysis of clinical tissue samples, with potential to advance research into disease mechanisms and the development of personalized medicine. In the future, the team plans to further optimize DynaTag to explore epigenetic regulatory mechanisms in more physiological and pathological processes.