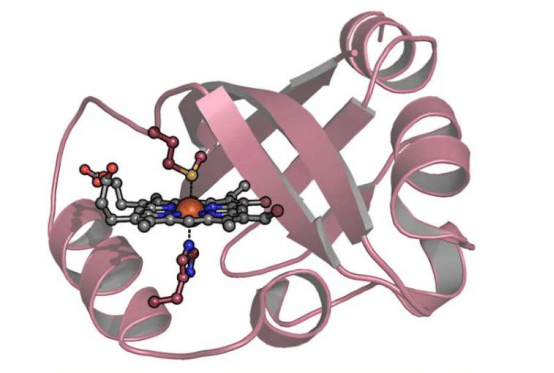
A research team led by Professor Joongoo Lee from the Department of Chemical Engineering at Pohang University of Science and Technology (POSTECH) has published groundbreaking work in Nature Communications, successfully using the ribosome to synthesize a variety of cyclic protein backbone structures. This discovery overcomes the billions-of-years-old limitation that ribosomes can only produce linear proteins, opening new avenues in biomedicine and materials science.
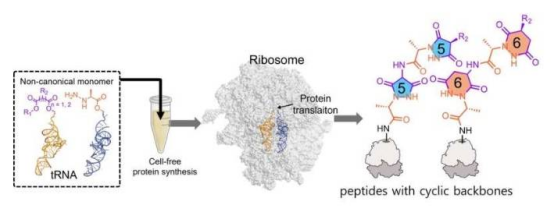
The team designed 26 special amino acids as novel building blocks that naturally interact inside the ribosome to form cyclic structures. Using a cell-free protein synthesis system, the researchers demonstrated that the ribosome can efficiently catalyze the formation of five- and six-membered cyclic backbones under physiological conditions (37°C, pH 7.5) without any external intervention. The formation efficiency of five-membered rings approached nearly 100%, while six-membered ring efficiency could be precisely tuned by adjusting substituents.
Professor Joongoo Lee stated: "What surprised us most is that the reaction mechanism inside the ribosome is so similar to conventional chemical processes." This finding not only expands the chemical space of biopolymers but also offers fresh insight into the primitive translation systems at the origin of life.
Key breakthroughs of the technology include:
First-ever ribosome-mediated synthesis of diverse cyclic backbones
Mild reaction conditions fully compatible with biological systems
Cyclization efficiency precisely controllable through molecular design
Lays the foundation for developing novel cyclic peptide therapeutics
The study builds on the team's 2022 work, when they first reported that ribosomes could synthesize proteins containing six-membered rings. The new research dramatically expands the range of synthesizable structures and proves that the ribosome can act as a versatile catalyst for non-canonical reactions.