A team led by molecular biologist Danny Incarnato at the University of Groningen in the Netherlands has published significant findings in Nature Biotechnology, identifying hundreds of RNA structural switches that regulate gene expression in E. coli and human cells. This study offers fresh perspectives on understanding gene regulation mechanisms.
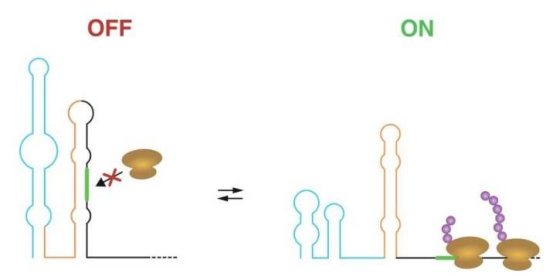
The team developed an innovative tool that combines evolutionary information to accurately identify functional RNA structural switches. Incarnato stated: "The ability of RNA to switch between alternative structures often indicates some form of regulatory mechanism." Experiments confirmed that these switches control ribosome binding to RNA through structural changes, thereby modulating protein translation.
One typical example revealed is a temperature-responsive switch in bacteria that helps cope with cold stress. By analyzing different conformations of RNA molecules in living cells, the researchers successfully mapped these regulatory switches in detail. Postdoctoral researcher Ivana Borovska noted: "Identifying these switches is only the first step; the next is to explore ways to regulate their function."
This three-year study builds on six years of foundational work and lays the groundwork for developing novel therapies targeting RNA switches. Future research will investigate how to design small molecules to precisely control these switches, potentially opening new avenues for disease treatment.