A research team from the University of Glasgow has developed a novel protein language model, PLM-Interact, capable of effectively predicting protein interactions and analyzing the impact of mutations. Published in Nature Communications, this study leverages supercomputing resources to provide a new tool for disease mechanism research and drug target discovery.
The team, led by Dr. Ke Yuan from the School of Cancer Sciences, Professor Craig Macdonald from the School of Computing Science, and Professor David L. Robertson from the Centre for Virus Research, developed the PLM-Interact protein language model based on large language models. Trained on over 421,000 human protein interaction pairs, the model demonstrates superior predictive performance compared to existing models.
Dr. Ke Yuan stated: "The DiRAC supercomputer, originally used to study the laws of nature, has now helped us build a new model for exploring protein interactions. Colleagues from the School of Computing Science provided language modeling support, while DiRAC's computational resources enabled us to complete this work more efficiently." The protein language model involves over 650 million parameters during training, accelerated by the GPU clusters of the UK's DiRAC high-performance computing facility.
PLM-Interact achieves 16% to 28% higher accuracy in predicting protein interactions compared to other advanced AI models. It successfully predicted protein interactions related to five key biological functions, while other tools could only predict one. The study also confirmed that the model accurately identifies the effects of mutations on protein interactions, including those causing hereditary diseases and cancer.
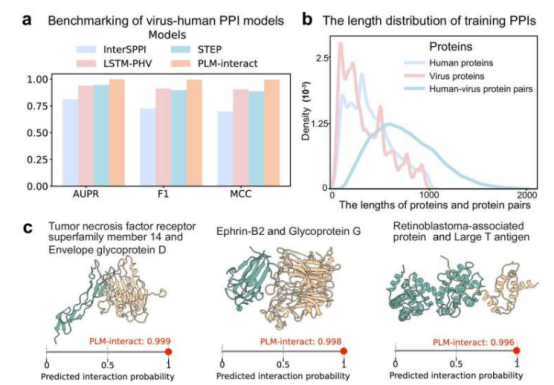
The researchers further trained the model using 22,383 interaction data points from human and viral proteins. PLM-Interact also performed excellently in predicting virus-host protein interactions, demonstrating its potential in virology research. Professor David L. Robertson noted: "The COVID-19 pandemic highlighted the urgency of understanding virus-host interactions. Tools like PLM-Interact can help us better understand viral emergence and disease risk."
The development of this protein language model provides a new platform for large-scale, precise prediction of protein interactions and is expected to play a significant role in future disease mechanism research and therapeutic development.