A research team from the Max Planck Institute of Biology, the University of Tübingen, and the University Hospital has recently developed an innovative protein design method, providing new approaches for targeted drug development. The research findings have been published in the journal Advanced Science.
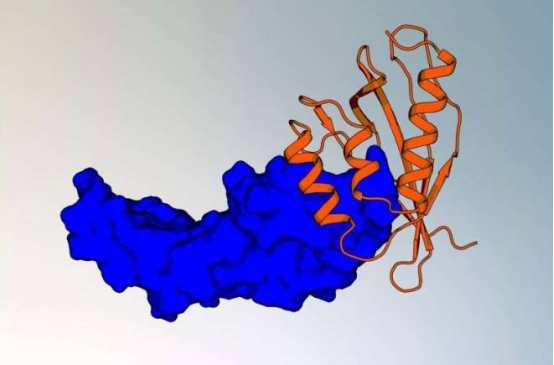
The computational workflow developed by the team uses complementary shape matching technology to design protein binders without relying on neural network training. First author Kateryna Maksymenko stated: "Our goal is to develop a training-free design workflow that not only creates site-specific binders but also enhances understanding of protein functions."
The method has been successfully applied to design protein binders targeting two key biomolecules: IL-7Rα, associated with immune function and leukemia, and VEGF, involved in angiogenesis. By integrating protein database screening, physical interface design, and molecular dynamics simulations, the team developed binders with high affinity and stability.
"This design workflow simplifies the protein engineering process while deepening our understanding of the physical basis of protein functions," the researchers noted. The new method can also incorporate non-canonical amino acids into designed proteins, offering possibilities for developing safer therapeutic drugs.
The technology is entirely based on first principles and is applicable not only to genetically encodable proteins but also to synthetic protein design. The research team stated that this breakthrough is expected to accelerate therapeutic drug discovery and bring new opportunities for molecular diagnostics.