A University of Pennsylvania research team has published groundbreaking findings in Nature Communications, using deep learning to screen animal venom for potential antibiotic molecules—a novel approach to combating the growing crisis of antibiotic resistance.
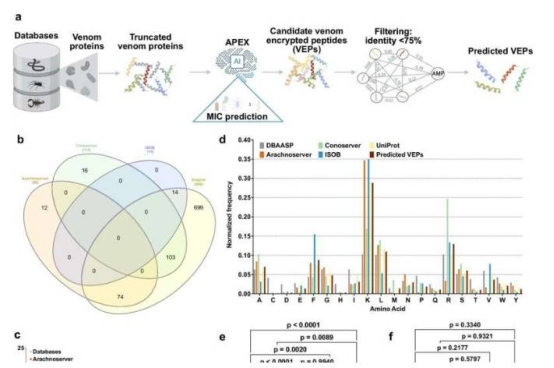
The researchers developed an artificial intelligence system called APEX that rapidly screened over 40 million venom-encrypted peptides (VEPs). These tiny proteins, produced by snakes, scorpions, spiders, and other venomous organisms, have evolved unique antibacterial properties. In just hours, APEX identified 386 candidate molecules with strong antibiotic potential.
"Venom is a masterpiece of evolution, yet its antibacterial potential has been largely unexplored," said lead researcher Dr. César de la Fuente. The team then synthesized 58 of these candidates for laboratory testing and found that 53 effectively killed drug-resistant Escherichia coli and Staphylococcus aureus while showing no toxicity to human red blood cells.
The study also uncovered more than 2,000 previously unknown antimicrobial motifs—short amino acid sequences critical to the antibacterial function of peptides. The team is now structurally optimizing the most promising candidates with the goal of developing a new generation of antibiotics.